3D Balaban index¶
The geometric distance matrix is defined as
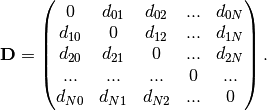
and leads to the adjacency matrix 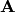 that contains the elements
that contains the elements
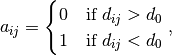
where 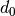 is a threshold that defines the existence of a bond. Similar to the Wiener number, the 3D Balaban index
is a threshold that defines the existence of a bond. Similar to the Wiener number, the 3D Balaban index
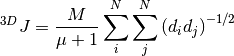
represents a topographical descriptor related to the shape of the molecule. Here, 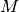 is the number of edges in the molecule,
is the number of edges in the molecule, 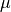 is the cyclomatic number
is the cyclomatic number
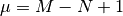
and 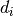 is the sum of all entries in the
is the sum of all entries in the 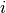 -th row of the
-th row of the 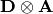 matrix.
matrix.
The use of the Balaban index is invoked by the following block in the meta-config.json file:
"collective variables": [
{
"type": "balaban",
"name": "2d balaban index",
"exclude_h": False
"update_adjacency": False
},
...
]
The additional keyword "exclude_h" allows for the calculation of the distance matrix without considering distances from and to Hydrogen atoms. If "update_adjacency" is set to True, the
adjacency matrix is recalculated in every dynamics step and therefore accounts for changes in the molecular connectivity.